At this time, interest in virus analysis using high-throughput, multi-parameter techniques such as flow cytometry are of interest. The use of flow cytometry for virus samples are similar to those of other nano-sized biological particles such as extracellular vesicles. The use of flow cytometry for nanoparticles is non-trivial and a number of publications and initiatives have been undertaken to address the lack of standardization of nanosized sample analysis as well as identifying and optimized the sensitivity of flow cytometry equipment. The translational nanobiology section has been involved in the development of nanoparticle flow cytometry analysis and sorting assays as well as initiatives to Improve cross-institution and platform standardization.
Flow Cytometry of single viral particles | Analysis & sorting

SARS-CoV2 is very similar in size to HIV, a retrovirus that we used in collaboration with NCI Vaccine Branch scientists to optimize and demonstrate a method for flow cytometric analysis and sorting of functional viral particles 1-3. Careful instrument configuration and sample processing is required to ensure accurate interpretation of results from studies such as these, since these viruses are very small and near the limits of detection for even highly specialized flow cytometers. All such studies should be carefully designed and reported in a manner that complies with the ISEV-ISAC-ISTH MIFlowCyt-EV Reporting Framework 4.
Flow Cytometry of single viral particles | Calibration
The use of calibration is critical when analyzing small particles due variations in instrument sensitivity5. Without standardization of data comparisons cannot easily be made (Figure 1A, B). The calibration of data not only allow instrument to instrument comparisons but can also enable better characterization of the particles of interest e.g. deriving size or the number of copies of protein on their surface (Figure 1E, F).
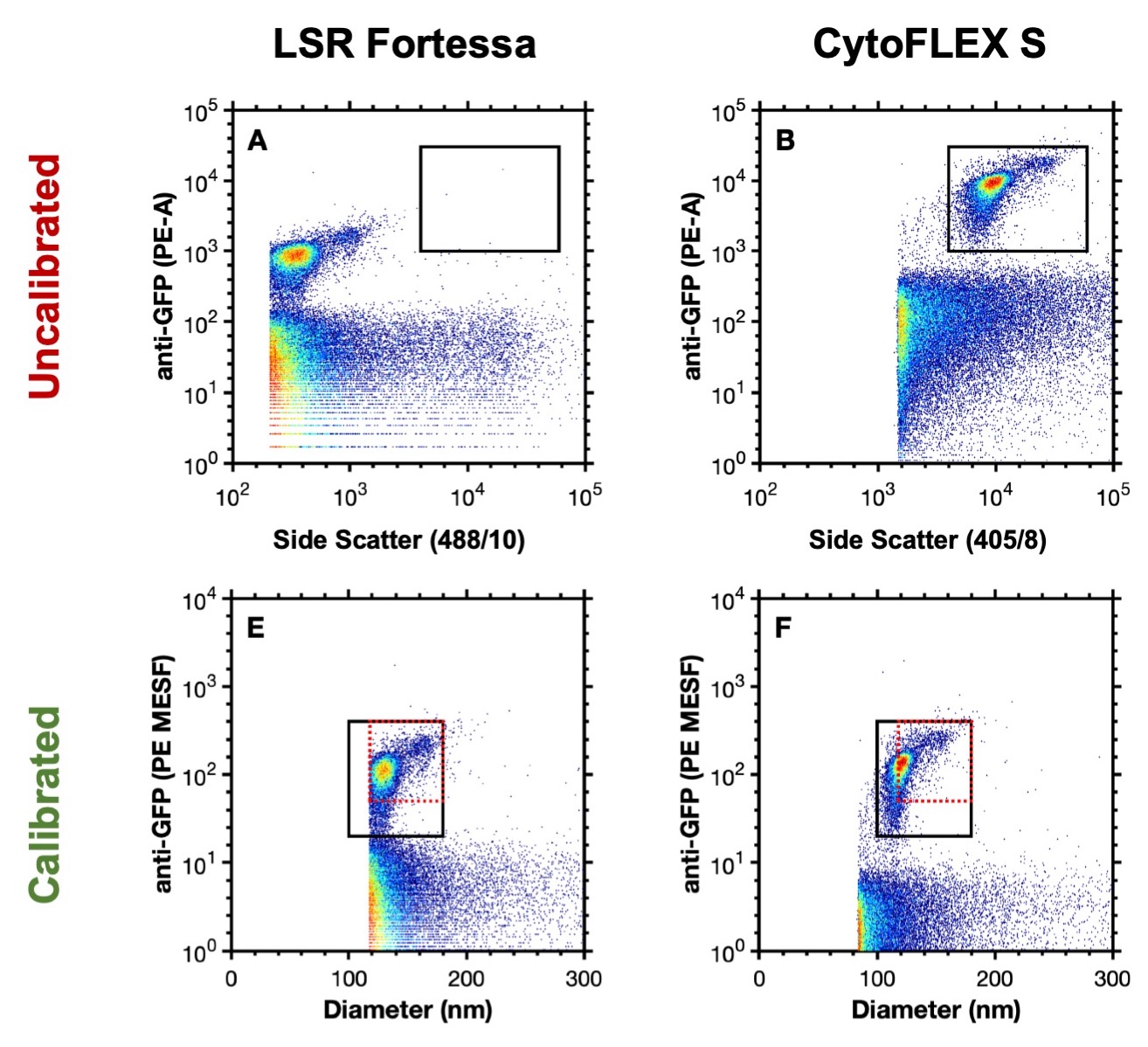
Figure 1. Uncalibrated data analyzing murine leukaemia virus (124 nm) on an LSR Fortessa (A) and CytoFLEX S (B). Calibrated data analyzing murine leukaemia virus on an LSR Fortessa (E) and CytoFLEX S (F). Data from Welsh J A, Jones J.C. Tang V A., Fluorescence and light scatter calibration allow comparisons of small particle data in standard units across different flow cytometry platforms and detector settings., Cytometry Part A, doi: 10.1002/cyto.a.24029.
Along with analysis, the sorting of intact biological nanoparticles (such as viruses) is also a potential useful technique for their functional characterization. The translational nanobiology section has developed and validated sorting techniques using the HIV virus (Figure 2A-C) and demonstrated is purity through re-analysis, along with functional activity (Figure 2D)3.

Figure 2. (a) Representative dot plots of PBS and mixed virus scattering. (b) Presort dot plots of PBS and mixed virus (top), post-sort reanalysis of NL4.3 virus and Bal virus (bottom). (c) Summary of gate quadrant events from panel (b) highlighting sort efficiency, lack of coincidence detection, and percentage purity. Red boxes highlight system noise. (d) Shows electron micrographs with representative pre- (top left) and post- (bottom left) nanoFACS sorted viral material, demonstrating characteristic viral features (immature and mature virus particles are visible, as well as well-defined cone-shaped core structures). Microscopy shows that nanoFACS sorted virus from panel (a,b) specifically infects only the cell line expressing the appropriate co-receptor for the respective sorted viral population. HIV-1 Bal (CCR5-tropic) stained with PKH26 and HIV-1 NL4.3 (CXCR4-tropic) stained with PKH67 were sorted apart from one another and titred on reporter U373-MAGI-CCR5 and U373-MAGI-CXCR4 cells, which show β-Gal (blue) staining when infected (arrows). Sorted CCR5-tropic Bal viral particles infect cells expressing CCR5 (top middle), but not cells expressing CXCR4 (bottom middle). Sorted NL4.3 does not infect cells expressing CCR5 (top right) but does infect cells expressing CXCR4 (bottom right). Data from Aizea Morales-Kastresana, Thomas A. Musich, Joshua A. Welsh, William Telford, Thorsten Demberg, James C. S. Wood, Marty Bigos, Carley D. Ross, Aliaksander Kachynski, Alan Dean, Edward J. Felton, Jonathan Van Dyke, John Tigges, Vasilis Toxavidis, David R. Parks, W. Roy Overton, Aparna H. Kesarwala, Gordon J. Freeman, Ariel Rosner, Stephen P. Perfetto, Lise Pasquet, Masaki Terabe, Katherine McKinnon, Veena Kapoor, Jane B. Trepel, Anu Puri, Hisataka Kobayashi, Bryant Yung, Xiaoyuan Chen, Peter Guion, Peter Choyke, Susan J. Knox, Ionita Ghiran, Marjorie Robert-Guroff, Jay A. Berzofsky & Jennifer C. Jones (2019) High-fidelity detection and sorting of nanoscale vesicles in viral disease and cancer., Journal of Extracellular Vesicles, 8:1, DOI: 10.1080/20013078.2019.1597603.
Translational Nanobiology Section Resources
Here you can find resources for understanding and implementing nanoparticle flow cytometry. These include:
- References to a selection of articles useful for getting started in the nanoparticle flow cytometry field, as well as detailed articles addressing light scatter and fluorescence calibration, and field position papers.
- A background to flow cytometry light scatter optics.
- Software developed by the translational nanobiology section for the calibration of flow cytometry data.
References
- Musich, T.; Jones, J. C.; Keele, B. F.; Jenkins, L. M. M.; Demberg, T.; Uldrick, T. S.; Yarchoan, R.; Robert-Guroff, M., Flow virometric sorting and analysis of HIV quasispecies from plasma. JCI Insight 2017, 2 (4). doi: 10.1172/jci.insight.90626. [website]
- Morales-Kastresana, A.; Telford, B.; Musich, T. A.; McKinnon, K.; Clayborne, C.; Braig, Z.; Rosner, A.; Demberg, T.; Watson, D. C.; Karpova, T. S.; Freeman, G. J.; DeKruyff, R. H.; Pavlakis, G. N.; Terabe, M.; Robert-Guroff, M.; Berzofsky, J. A.; Jones, J. C., Labeling Extracellular Vesicles for Nanoscale Flow Cytometry. Sci Rep 2017, 7 (1), 1878. doi: 10.1038/s41598-017-01731-2. [website]
- Morales-Kastresana A*, Musich T*, Welsh J A*, Telford W, Demberg T, Wood J C S, Felton E J, Bigos M, Ross C D, Kachynski A, Dean A, Dyke J V, Tigges J, Toxavidis V, Parks D R, Overton W R, Kesarwala A P, Freeman G J, Rosner A, Perfetto S P, Pasquet L, Terabe M, McKinnon K, Kapoor V, Trepel J B, Puri A, Kobayashi H, Yung B, Chen X, Guion P, Choyke P, Knox S J, Ghiran I, Robert-Guroff M, Berzofsky J A, Jones J C, High Fidelity Detection and Sorting of Nanoscale Vesicles in Viral Disease and Cancer , Journal of Extracellular Vesicles, doi: 10.1080/20013078.2019.1597603. [website]
- Welsh J A, van der Pol E, Arkesteijn G, Bremer M, Brisson A, Coumans F, Dignat-George F, Duggan E, Ghiran I, Giebel B, Görgens A, Hendrix A, Lacroix R, Lannigan J, Libregts S, Lozano-Andrés E, Morales-Kastresana A, Robert S, de Rond L, Tertel T, Tigges J, de Wever O, Yan X, Nieuwland R, Wauben M, Nolan J, Jones, J C., MIFlowCyt-EV: a framework for standardized reporting of extracellular vesicle flow cytometry experiments, Journal of Extracellular Vesicles. doi: 10.1080/20013078.2020.1713526. [website]
- Welsh J A, Jones J.C. Tang V A., Fluorescence and light scatter calibration allow comparisons of small particle data in standard units across different flow cytometry platforms and detector settings, Cytometry Part A, doi: 10.1002/cyto.a.24029. [website] [cytometry files - 720 MB]